Tests for epistasis of bimD with UV repair genes representing four epistatic groups and their putative types of DNA repair in Aspergillus nidulans
Etta Kafer. Department of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, B.C., V5A 1S6, Canada
bimD6, a ts mitotic mutant sensitive to UV only in dividing cells and defective in recombination, was tested for epistasis with members of the four Uvs groups of A. nidulans. UV survival of quiescent and germinating conidia was compared. While details varied between tests, results from UV-treated germinating conidia consistently showed additive or synergistic interaction. This suggested a new type of function for bimD and perhaps indirect effects on DNA repair and recombination, in line with recent evidence for defects in chromosome cohesion and compaction in mutants of bimD and its homologs (van Heemst et al. 2001 PNAS 98:6267-6272).______________________________________________________________________________
The ts mutant, bimD6, which is blocked in mitosis at 42°, was found to be meiotic-defective and hypersensitive to UV and MMS (methyl-methane sulfonate) during growth at low temperature. When cloned its sequence showed no homology to any known gene (Denison et al. 1992 Genetics 134:1085-1096). We therefore started a project testing bimD for epistasis with uvs gene members of the four Uvs groups identified in Aspergillus (Chae and Kafer 1993 Curr. Genet. 24:67-74). Some of these groups contain genes with mutants of very similar phenotypes, suggestive of functional grouping, as known for epistatic groups of yeast RAD genes involved in radiation repair. Similarities and differences of the Aspergillus Uvs groups compared to the budding yeast RAD groups and repair types are increasingly becoming documented (see below). In Aspergillus, conidia are dormant non-metabolizing spores, basically in G0, and the first mitosis occurs only after several hours of exposure to growth conditions. Certain repair-defective mutants are found to be UV sensitive when quiescent conidia are treated; presumably in such cases repair can occur in haploid non-dividing cells, as typically occurs in excision and possibly in other types of error-free repair. Alternatively, mutants may show sensitivity only when growing cells are treated, as was found for bimD6 and is typical for mutants defective in recombinational repair. The latter type of treatment was therefore more likely to give meaningful results but we used both methods, because synergism in double mutants has occasionally also been found when one or both of the single mutants did not show sensitivity (e.g., in MMS tests of uvsA101 and uvsI501, neither of which is hypersensitive to MMS, the double mutant showed high sensitivity; Chae and Kafer 1993 ref. cit.). Single and double mutant strains of bimD6 and uvs mutations from each of the four epistatic groups were compared for UV survival using parallel platings of UV treated conidia, irradiated either immediately (data not shown) or after preincubation (4 examples in Fig. 1). Overall, the results of tests using dividing cells showed additive or synergistic interaction of bimD6 with all of the tested mutations. However, in a few cases the difference between double and single mutants were barely additive; namely for uvsD153 (Fig. 1C), and especially for musN227 which is not UV sensitive (Fig. 1D). In addition, since for such tests null mutations were not available (especially for bimD and uvsF which are essential genes) these results have to be interpreted with caution. However, conclusions can be reached with some confidence when the null phenotype is known to be identical to that of the mutants used. This appears to be likely for uvsI501, and has recently been shown for the cell cycle checkpont genes uvsB and uvsD, after sequencing and construction of disruption alleles (De Souza et al. 1999 Mol. Biol. Cell 10:3661-3674). This is however not the case for musN227 which will need further testing. (musN227 had produced inconsistent results which could well have been due to partial function; namely, while musN227 apparently showed typical epistatic interaction with uvsC114 in tests for MMS survival, non-epistatic interaction, i.e., significantly increased UV-induced mutation was obtained for the musN227;uvsC114 double mutant strain; Kafer and Chae 1994 Curr. Genet. 25:223-232; Chae 1993 Ph.D. Thesis, McGill Univ. Montreal Canada). In fact, the original musN227 mutant was recently shown to differ from musN? types which have poor growth and high sensitivity to MMS and UV, in line with the identified homology of musN to E. coli, fungal and human recQ helicases and their extreme mutant phenotypes (A. Hofmann, personal communication). In several cases, the findings after pregermination were confirmed by at least additive interaction also after treatment of quiescent conidia; namely, for uvsF201 and uvsI501 results were almost identical (shown for uvsI in Fig. 1 A) and for uvsB110 the double mutant with bimD6 showed synergism in either test but considerably more after pregermination. On the other hand, for mutants with dormant conidia that are not UV sensitive, i.e., uvsE182, a recombination-deficient type similar to the recA homolog uvsC114, and also for musN22, treatment of quiescent spores produced overlapping survival curves of all mutant and control strains. Such results differed clearly from those obtained after pregermination (Figs 1B and 1D). The overall conclusion, therefore, is that bimD most likely is not a member of one of the four identified epistasis groups of UV repair genes in Aspergillus. This agrees with recent findings which suggest a different primary function for bimD from that proposed for genes of the four Uvs groups (see below). While genetic analysis of bimD6 showed extreme reduction of spontaneous mitotic recombination in diploids, similar to that found for the recA homolog uvsC114, intrachromosomal conversion in bimD6 duplications was practically at wild type levels but almost absent in uvsC114 (van Heemst et al. ref. cit.). Furthermore, detailed cytoimmunological analysis localized the abundant BIMD protein on the chromosomes at most stages of the meiotic and mitotic cell cycle, as found also for its Sordaria homolog, SPO76 (van Heemst et al. 1999 Cell 98:261-271). It was therefore concluded that bimD is unlikely to be enzymatically involved in recombination, DNA repair or cell cycle progression, but rather functions in mitotic and meiotic chromosome morphogenesis, possibly affecting chromosome cohesion and compaction. This conclusion is supported by analysis of bimD6 suppressors (e.g., Holt and May 1996 Genetics 142:777-787) and also by results in yeast and human homologs (reviewed by van Heemst and Heyting 2000 Chromosoma 109:10-26). Such bimD function differs from that of members in the four Uvs epistatic groups of A. nidulans which increasingly is becoming better defined [superceding even recent reviews; e. g., Kafer and May 1998 In Nickoloff and Hoekstra (eds), DNA Damage and Repair, vol.1, Humana, Totowa NJ, p. 477-502]. Current analysis also reveals more clearly similarities and differences to the model provided by budding yeast. Namely, yeast RAD genes can be classified into three essentially non-overlapping groups by epistasis and by types of radiation-repair function: 1) RAD3 group, involved in nucleotide excision repair (NER; mutants mainly UV sensitive); 2) RAD6 group, active in error-free and mutagenic postreplication repair (mutants UV and X-ray sensitive); 3) RAD52 group, required for homologous recombination and double strand break repair (mutants mainly X-ray sensitive). While comparison of A. nidulans epistatic UV repair groups to yeast groups (2) and (3) reveals interesting partial correspondence, no information about group (1), i.e., excision repair, is available right now. The hypothesis is that in Aspergillus details of excision processes and mutants differ from those described for budding yeast, but correspond to the situation in Neurospora (and also in fission yeast); more specifically, that two types of excision repair are active, one being specific for UV dimers (Yajima et al. 1995 EMBO J 14:2393-2399) the other resembling yeast and human NER (Hatekayama et al. 1998 Curr. Genet. 33:276-283). Provided both processes can partially substitute for each other, only the double mutant, but neither type of single mutants is very UV sensitive, which accounts for the lack of analysis of NER genes in Aspergillus (even though some of them are known to exist and be active). Interpretation of current results with respect to post-replication repair, i.e., function of yeast RAD6 group genes, reveals fairly close correspondence with Aspergillus genes of two groups, "UvsF" and "uvsI". A) The "UvsF"or better "UvsH or UvsJ" epistatic group, includes so far three genes: a) uvsF, a homolog of human and yeast RFC1, is an essential gene and codes for the largest component of DNA replication factor C (Kafer and May 1997 Gene 191:155-159); uvsF is therefore is not specifically a repair gene. Apparently, a point mutation in uvsF201 interferes solely with its repair function (S-K Chae, personal communication) resulting in defects resembling those found for mutants in excision repair genes while retaining normal growth [similar cases have been reported for other mutated replisome components in yeast; e. g., pol30-46 is repair defective resulting from a point mutation in PCNA (=proliferating cell nuclear antigen) which interacts with RFC and is essential for DNA replication (Xiao et al. 2000 Genetics 155:1633-1641)]. b) The two other genes of this group, uvsH and uvsJ, are homologs of RAD18 and RAD6, the two major DNA repair genes of the yeast RAD6 group. Both genes, which interact, are required for important error-free as well as all mutagenic postreplication repair. uvsH mutants resemble closely their rad18 homologs (Yoon et al. 1995 Mol. Gen. Genet. 248:174-181) being highly UV and X-ray sensitive, with increases in spontaneous and UV-induced mutation. uvsJ1 and uvsJ? share a similar phenotype, but their resemblance with rad6 is less close (Chae 1993 ref. cit. and personal communication). B) uvsI, the gene that defines the "UvsI" group, is a homolog of REV3 (Han et al. 1998 FEMS Microbiol. Lett. 164:13-19) which codes for the catalytic subunit of the mutagenic bypass DNA polymerase z . REV3 belongs to a RAD6 subgroup and equivalent function of Aspergillus uvsI seems likely. Such primary polymerase function had indeed been postulated first based purely on genetic analysis of mutation specificity, after uvsI501 was found to be defective in reversion only of certain types of point mutations (Chae 1993, ref. cit.; Chae and Kafer 1997 Mol. Gen. Genet. 254:643-653). C) Similarly, there is good correspondence for recombinational repair, i.e., between genes of the yeast RAD52 group and genes of the Aspergillus "UvsC" group. The latter includes uvsC, a recA/RAD51 homolog (van Heemst et al. 1997 Mol.Gen. Genet. 254:654664) which interacts with radC, a homolog of RAD52 (S-K.Chae; personal communication). Mutants of both genes are especially sensitive in growing cells, to MMS and UV rather than X-ray, and neither disruption is lethal. Two further genes, uvsE and musL, have similar rec- mutant phenotypes. All these Aspergillus genes are also meiotic defective and show increased chromosome malsegregation, similar to yeast rad52 and mutants of other genes of the RAD52 group. D) The two genes in the fourth epistatic Aspergillus group, "UvsB", have been identified as homologs to fission yeast cell cycle check-point control genes, uvsB showing homology to rad3 and the related human ATM/ATR genes, and uvsD to rad26 (De Souza et al. 1999 ref cit.). These findings explain their unusual features which differ not only from mutants of known yeast UV repair genes, but also from mutants of the budding yeast homologs, MEC1 and PIE1. These latter genes, but not those of fission yeast or Aspergillus, are essential genes presumably with additional functions (Desany et al. 2001 Genes Dev. 12:2956-2970; Wakayama et al. 2001 Mol. Cell. Biol. 21:755-764). Both, the Aspergillus uvsB and uvsD mutants, also show high levels of genomic instabilities. However, since mitotic levels of intra- and intergenic homologous recombination are clearly increased, such instabilities are not likely to result from defects in recombination, as found for rec- mutants of Aspergillus uvsC and uvsE (Jansen 1970 Mutat. Res. 10:33-41; Fortuin 1971 Mutat. Res. 11:265-277; Kafer and Mayor 1986 Mutat. Res. 161:119-134) and also is known for Rad51 homo- and paralogs in a variety of organisms (e. g., Takata et al. 2001 Mol. Cell. Biol. 21:2858-2866). Acknowledgement: The diligent assistance by Tania Fan with the various UV sensitivity tests is very much appreciated.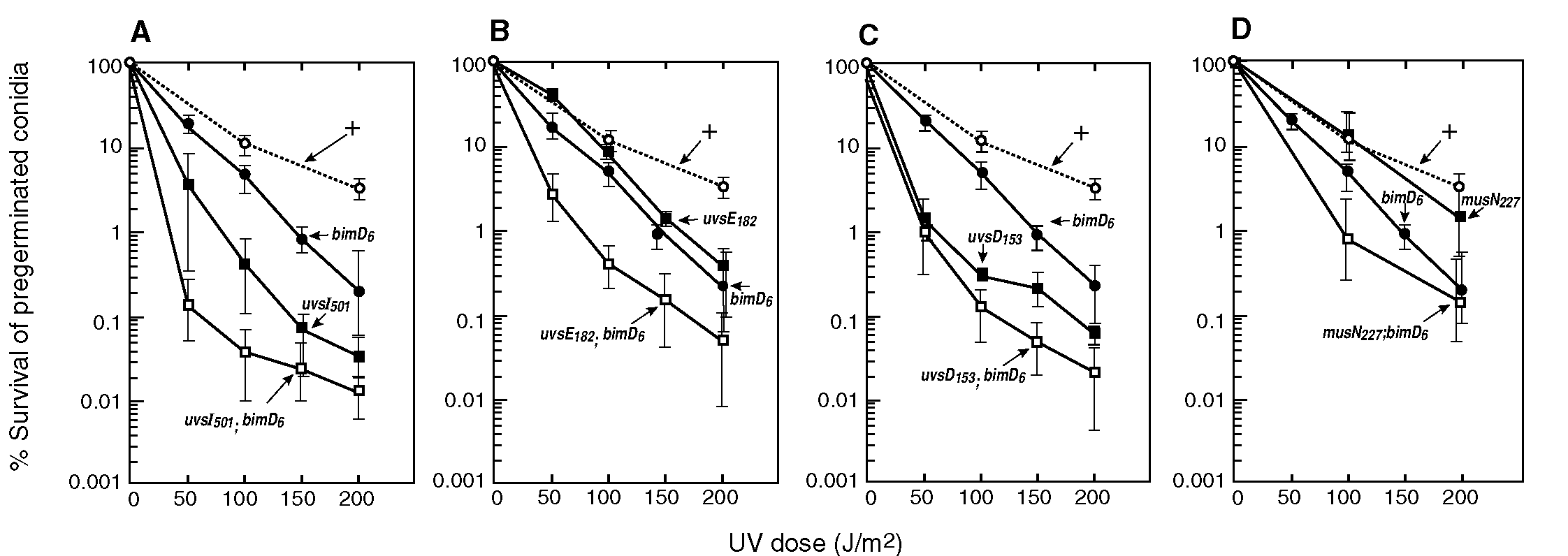 Fig. 1: UV survival of bimD6 and various UV-sensitive mutants, compared to their congenic double mutant strains
(as indicated) and to the repair-proficient (+) control. Conidia were plated, pregerminated by incubation at 30° for 5 1/2 hours,
and irradiated on the plates for various times up to a UV dose of 200 J/m2, using a dose rate of 1.6 J/m2/s.
Click HERE for a larger version of this figure
Fig. 1: UV survival of bimD6 and various UV-sensitive mutants, compared to their congenic double mutant strains
(as indicated) and to the repair-proficient (+) control. Conidia were plated, pregerminated by incubation at 30° for 5 1/2 hours,
and irradiated on the plates for various times up to a UV dose of 200 J/m2, using a dose rate of 1.6 J/m2/s.
Click HERE for a larger version of this figure