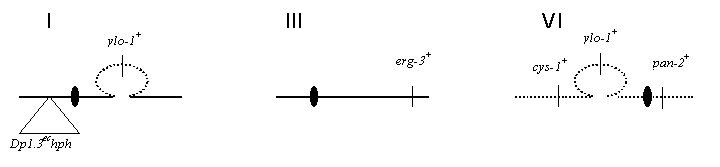
Figure 1.: Relative map positions in the Dp 1.3ec hph ; Dp(IBj5) mat a strain.
______________________________________________________________________________
Neurospora strains bearing large (e.g., >100 kb) chromosomal segment duplications (segmental aneuploids) can be obtained as segregants from crosses heterozygous for some insertional or quasiterminal translocations (reviewed in Perkins 1997 Adv. Genet. 36: 239-398). Sexual crosses made with segmental aneuploid strains display a characteristic "barren" phenotype; they form perithecia but only a few viable ascospores. Although the barren phenotype is useful in signalling the presence of segmental aneuploidy, it makes subsequent genetic analyses very tedious because of the difficulty in obtaining sufficient numbers of progeny. Therefore a need was felt to identify euploid strains that might increase ascospore yields in crosses with segmental aneuploid strains. A recent study involving crosses between Dp(AR17) duplication strains and eight wild-isolated strains revealed that crosses with two wild strains, Golikro (FGSC# 4830) and Costa Rica (FGSC# 852), produced significantly (approx 100x) fewer ascospores (Noubissi et al. 2000 Fungal Genet. Biol. 31: 91-97). Results summarized in Table 1 show this is true even in crosses with two other duplication strains, Dp(OY329) and Dp(S1229) and suggested that the productivity of barren crosses can be influenced by the euploid parent. This motivated us to examine wild-isolated strains for any that increased ascospore production in otherwise barren crosses. We examined 71 of the 74 wild-isolated N. crassa strains listed as mat A in the FGSC strain catalogue (7th edition, 1998). The three strains not tested were Lankala Koderu-1, FGSC 3358, Roanoke-1m, FGSC 2227; and Libreville, FGSC 4823. Each wild strain was crossed with Dp(AR17), Dp(OY329) and Dp(S1229) strains. The Dp(S1229) strains FGSC #264 and FGSC #265 were obtained from the FGSC. The Dp(AR17) strains A30 and A40 , and the Dp(OY329) strain C25-3 are described by Bhat and Kasbekar (2001 Genetics 157: 1581-1590). Another Dp(OY329) strain, C23-13, was constructed by a similar approach to that used for constructing C25-3. The crosses were made on synthetic crossing medium in petri dishes and after 31 days the ascospore yields were estimated by inspecting the lids in a dissection microsope. Discernibly more ascospores were produced in the crosses with the wild-isolates Lahore-1 (FGSC #1824), Dagguluru-1 (FGSC #3360), Okeechobee (FGSC # 3968) and Tiassale (FGSC # 4825). Dp(AR17), Dp(OY329) and Dp(S1229) have previously been shown to suppress RIP in a smaller gene sized duplication, presumably by titrating out the RIP machinery (Bhat and Kasbekar 2001 Genetics 157: 1581-1590). We verified that the Lahore-1, Dagguluru-1, Okeechobee and Tiassale strains did not interfere with the ability of Dp(AR17) and Dp(OY329) to effect such suppression (data not shown). Dp(IBj5) suppresses RIP in Dp1.3ec hph: We wanted to test whether Dp(IBj5) also suppresses RIP in a small duplication. Dp(IBj5) is an insertional duplication on IR of a VIL segment (from cpc-1 through ylo-1) and strains bearing this duplication can be obtained as segregants from crosses of the translocation strain T(VIL > IR) IBj5 cpc-1 mat A (FGSC #443) with normal sequence strains. The small duplication was represented by Dp 1.3 echph, which is comprised of a 1.3 kb fragment of erg-3 that is marked with the hph gene for hygromycin-resistance, and targets RIP to the erg-3 locus on LGIIIR (Prakash et al., 1999 Microbiology 145: 1443-1451). Colonies generated from erg-3 mutant ascospores have a characteristic morphology on Vogel's-sorbose agar medium and are easily scorable under a dissection microscope (Noubissi et al. 2000 Fungal Genet. Biol. 31: 91-97). The IBj5 translocation strain was crossed with a Dp 1.3ec hph strain to generate Dp 1.3ec hph ; Dp(IBj5) segregants. Crosses of these segregants with the standard laboratory wild type strains 74-OR23-1 mat A or OR8-1 mat a were so severely barren that despite repeated attempts we could not obtain sufficient numbers of progeny to test whether Dp(IBj5) suppresses RIP in Dp 1.3ec hph. Therefore we resorted to crossing a Dp 1.3ec hph ; Dp(IBj5) mat a segregant with the Lahore-1, Dagguluru-1, Okeechobee and Tiassale strains. In a single attempt we recovered, respectively, 182, >2500, 81 and 34 progeny from these crosses. None of these progeny were mutant in erg-3. Combining the results for the crosses with the Lahore-1, Okeechobee and Tiassale strains, the erg-3 mutation frequency is estimated to be less than 1/297 (i.e., < 0.3%). We examined 1003 progeny from the cross with Dagguluru-1, thus the erg-3 mutation frequency in this cross was less than 1/1003 (i.e., < 0.1%). These values are much lower than the erg-3 mutation frequencies in crosses of these four wild-isolates with a Dp 1.3ec hph mat a strain (Noubissi et al. 2000 Fungal Genet. Biol. 31: 91-97) and therefore allow us to conclude that the IBj5 duplication, like the AR17, OY329 and S1229 duplications, suppresses RIP in the smaller duplication Dp 1.3ec hph. AB was supported by a CSIR-UGC Junior Research Fellowship, FKN by the Third World Organization of Women in Science.

Figure 1.: Relative map positions in the Dp 1.3ec hph ; Dp(IBj5) mat a
strain.
Table 1 : Yield of ascospores from crosses of mat a segmental aneuploid strains Dp(S1229 ) and Dp(OY329 ) with selected wild isolates
|
Segmental Aneuploid strains |
Wild Isolates |
|||||||
|
"Low-RIP" strains |
"High-RIP" strains |
|||||||
|
|
Dacca |
Carrefour Dufort |
Adiopodoume |
Golikro |
ravesnwood-1 |
Agudas Rd-1 |
Costa Rica |
Merger |
|
Dp(S1229) |
6.6x104 |
5x105 |
8.3x104 |
19 |
105 |
1.5x105 |
20 |
1.5x105 |
|
Dp(OY329) |
5x104 |
5x104 |
82 |
19 |
2.5x104 |
20 |
78 |
2.5x104 |